Package tools#
Scipy implements a number of algorithms for calcualting the eigendecomposition:
where
\(V\) is the matrix whose columns are the eigenvectors
\(\Lambda\) is a diagonal matrix with the eigenvalues on the diagonal
Variants only calculate the eigenvalues (faster) or take advantage of symmetries / sparsity, and numerical stability.
Method |
Type |
Use Case |
|---|---|---|
|
Dense (general matrix) |
Computes all eigenvalues and eigenvectors of a square matrix. |
|
Dense (general matrix) |
Computes only eigenvalues of a square matrix (faster than |
|
Dense (symmetric/Hermitian) |
Computes eigenvalues and eigenvectors for symmetric or Hermitian matrices (numerically stable). |
|
Dense (symmetric/Hermitian) |
Computes only eigenvalues of symmetric or Hermitian matrices (faster than |
|
Sparse (symmetric/Hermitian) |
Computes a subset of eigenvalues and eigenvectors for sparse symmetric or Hermitian matrices. |
|
Sparse (general matrix) |
Computes a subset of eigenvalues and eigenvectors for sparse general (non-symmetric) matrices. |
Eigendecomposition is sometimes called diagonalization.
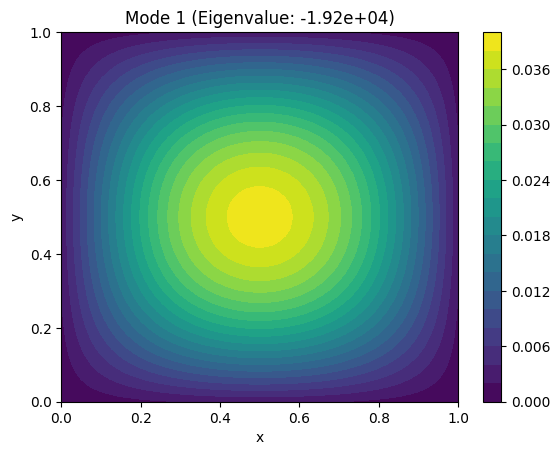
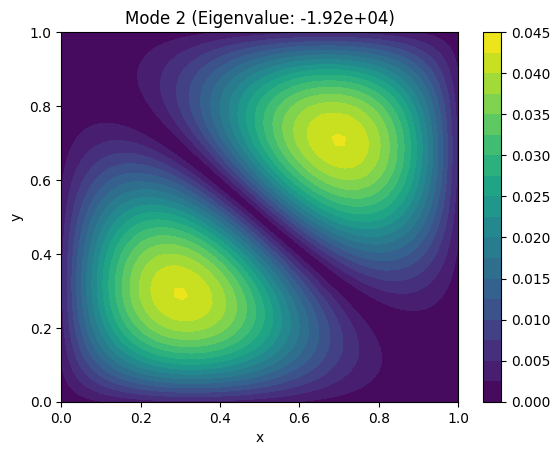
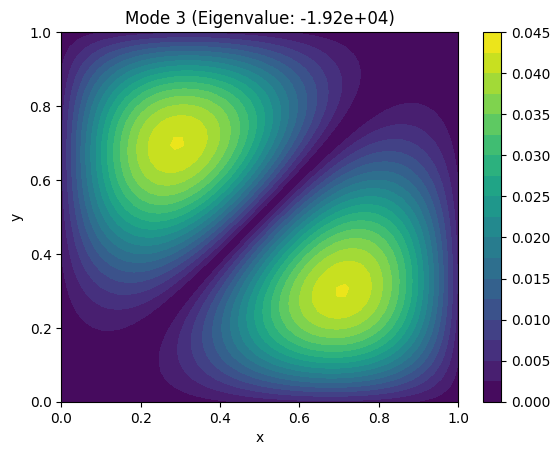
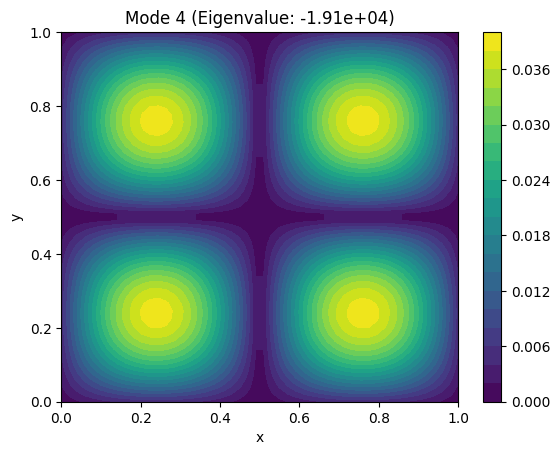
Example: Find the fundamental vibrational modes of a square membrane.#
The vibrational modes are solutions to the Laplace equation: $\( \nabla^2 u = 0 \quad u=0 \ on \ boundary\)$
import numpy as np
import matplotlib.pyplot as plt
from scipy.linalg import eigh
# Grid properties
Lx = 1.0 # Length in the x-direction
Ly = 1.0 # Length in the y-direction
Nx = 50 # Number of grid points in the x-direction
Ny = 50 # Number of grid points in the y-direction
dx = Lx / (Nx - 1) # Grid spacing in the x-direction
dy = Ly / (Ny - 1) # Grid spacing in the y-direction
# Construct the Laplacian operator for the 2D grid
def construct_laplacian_2d(Nx, Ny, dx, dy):
N = Nx * Ny
laplacian = np.zeros((N, N))
for i in range(N):
laplacian[i, i] = -2 / dx**2 - 2 / dy**2
if i % Nx != 0: # Left neighbor
laplacian[i, i - 1] = 1 / dx**2
if (i + 1) % Nx != 0: # Right neighbor
laplacian[i, i + 1] = 1 / dx**2
if i >= Nx: # Top neighbor
laplacian[i, i - Nx] = 1 / dy**2
if i < N - Nx: # Bottom neighbor
laplacian[i, i + Nx] = 1 / dy**2
return laplacian
# Construct the Laplacian matrix
laplacian = construct_laplacian_2d(Nx, Ny, dx, dy)
# Solve the eigenvalue problem
eigenvalues, eigenvectors = eigh(laplacian)
# Extract the first few modes
modes = 4
eigenfunctions = [eigenvectors[:, i].reshape(Nx, Ny) for i in range(modes)]
# Plot the first few eigenfunctions (vibration modes)
for i, eigenfunction in enumerate(eigenfunctions):
plt.figure()
plt.contourf(np.linspace(0, Lx, Nx), np.linspace(0, Ly, Ny), abs(eigenfunction), 20, cmap="viridis")
plt.colorbar()
plt.title(f"Mode {i+1} (Eigenvalue: {eigenvalues[i]:.2e})")
plt.xlabel("x")
plt.ylabel("y")
plt.show()